SITVIT WEB, online consultation of the WorldWide database of M. tuberculosis.
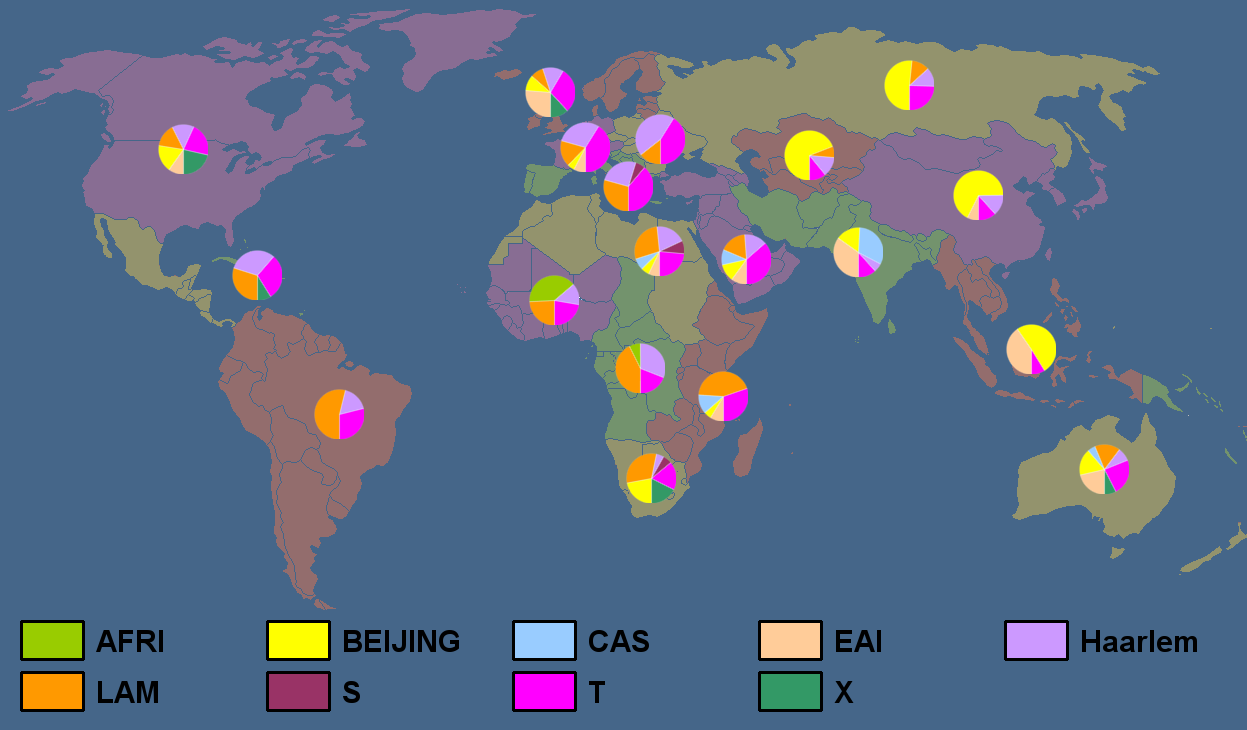
SITVIT is a Mycobacterium tuberculosis molecular markers database. It contains three major types of molecular markers: Spoligotypes, MIRU and VNTR markers. Spoligotypes are patterns formed by the presence or absence of 43 spacers in the DR locus of Mycobacterium tuberculosis. This pattern can be seen as a 43 digit pattern or under a reduced octal form of only 15 numbers. MIRU markers (Mycobacterial interspersed repetitive units) are variable number of tandem repeats markers. The MIRU method currently used on Mycobacterium tuberculosis is composed of 12 different MIRU loci. A mirutype is a 12 digits pattern representing the number of repetitions of each of these 12 specific loci. But Mycobacteria genome counts also several short nucleotide sequence organism as tadem repeat named Variable Number Tandem Repeats (VNTR). Five exact tandem repeat (ETR) are used for VNTR analysis of M. tuberculosis complex. This is the third marker contained in our database. Each ETR locus contains multiple tandem copies of a particular repeat unit, association of all of them generates alleles profile useful for the identification and the evolutionary studies of M tuberculosis complex.
Quantitative description of SITVIT WEB
This table is a description of SITVIT WEB database. More details in supplemental materials of the publication.
| Genotyping marker | Number of patterns | Number of strains | Number of IT | Number of ITed strains | Number of orphan strains | Percentage of orphan strains | Number of countries | |
|---|---|---|---|---|---|---|---|---|
| spoligotype | 7 104 | 58 187 | 2 747 | 53 830 | 4 357 | 7.5 | 102 | 105 |
| MIRU12 | 2 380 | 8 161 | 847 | 6 628 | 1 533 | 18.8 | 35 | |
| VNTR | 458 | 4 626 | 245 | 4 413 | 213 | 4.6 | 22 | |
Contributors
Following contributors kindly provided genotyping data to successive public databases SpolDB1 to SpolDB4 and the SITVITWEB projects; note that the names are given in order of the size of data provided from highest to smallest numbers of strains (only shown for 100 or more isolates): J. R. Driscoll, B. Kreiswirth, T. Victor et al.*, P. M. Hawkey, R. A. Skuce, N. Rastogi et al.**, P. Suffys, S. A. M. Sahal, L. Rigouts, N. Haddad, H. Soini, W. M. Prodinger, A. Gori, M. Fauville-Dufaux et al.***, T. N. Quitugua, U. Singh, C. Garzelli, D. van Soolingen et al.**** , V. Katalinic, S. Cheong, N. Morcillo, L. Aristimuno, L. Anh, M. C. Gutierrez, Z. Rahim, T. Koivula, S. Eyangoh, A. Karboul, S. Niemann, F. Stauffer, I. Mokrousov, M.J. Zumarraga, N. Kurepina, J. Dale, T. Lillebaeck, R. Jou, M. L. Rossetti, F. Chaves, H. Traore, S. David, D. Garcia de Viedma, I. Parwati, Y. J. Sun, A. Kwara, A. G. Van der Zanden, R. Durmaz, A. Sajduda, R. Hazan, V. Rasolofo-Razanamparany, A. Gibson, B. de Jong, C. Gilpin, M. P. Nicol, J. Maugein, D. Nguyen, R. Diaz, S. Gagneux, T. Iwamoto, C. A. Rodriguez, D. Alland, D. Cousins, V. Ritacco, G. Kallenius, P. Supply, S. Kulkarni, J. Martin, S. Panaiotov, M. H. Féres Saad, F.P. Sweeney, J. Bauer, L. S. Cowan, M. Goyal, M. Kubin, L. Binder, E. Kassa-Kelembho, J. T. Douglas, S. Bonora, O. Dellagostin, T. Tracevska, D. Almeida, O. Ogarkov, E. Aktas, R. Ohata, G. S. Kibiki, , P. Farnia, A. Michel, I. Shemyakin, M. Doroudchi, C. E. Salas, O. Kisa, C. Ellermeier, J. De Waard, V. Narbonne, Y. O. Goguet de la Salmonière, M. Ridell, G. Vrioni, F. Portaels.
* T. Victor contributed genotypes jointly with E. Streicher and R. M. Warren.
** N. Rastogi contributed genotypes jointly with his coworkers: A. Devallois, E. Legrand, C. Sola, L. Horgen, I. Filliol, S. Ferdinand, K. Brudey, T. Zozio, J. Millet, V. Hill, and D. Couvin.
*** M. Fauville-Dufaux contributed genotypes jointly with C. Allix
**** D. van Soolingen contributed genotypes jointly with K. Kremer.
Global Distribution of the strains recorded in SITVITWEB
Distribution of Clades in SITVITWEB